Golden Eagle Core Areas: Discriminant Analysis
Examine the table of means, it is apparent that the regions are very different with respect to certain variables. For example, region 1 has no wet heath and very little land below 200 m, however it does have a lot of bog.
| REGION | 1 (n = 7) | 2 (n = 16) | 3 (n = 17) | Total (n = 40) | ||||
|---|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | Mean | SD | Mean | SD | |
| POST | 1.7 | 1.52 | 0.9 | 2.44 | 2.7 | 2.95 | 1.8 | 2.63 |
| PRE | 3.7 | 1.76 | 0.8 | 2.10 | 2.0 | 3.03 | 1.8 | 2.64 |
| BOG | 13.2 | 2.61 | 4.5 | 2.77 | 8.7 | 3.82 | 7.8 | 4.46 |
| CALL | 0.8 | 0.76 | 2.0 | 2.38 | 2.9 | 2.10 | 2.2 | 2.16 |
| WET | 0.0 | 0.00 | 7.4 | 3.26 | 1.5 | 1.09 | 3.6 | 2.83 |
| STEEP | 4.4 | 1.39 | 9.3 | 5.02 | 1.9 | 0.78 | 5.3 | 4.70 |
| LT200 | 4.5 | 4.07 | 12.4 | 5.33 | 19.9 | 4.34 | 14.2 | 7.33 |
| L4_600 | 4.7 | 5.11 | 3.2 | 3.29 | 0.0 | 0.03 | 2.1 | 3.43 |
Discriminant functions
When there are more than 2 groups it may be possible to construct more than one discriminant function. Indeed the maximum number of discriminant functions that can be obtained is the lesser of:
- the number of groups - 1
- the number of predictor variables
Since there are 3 groups and 8 variables the maximum number of discriminant functions is 2.
Summary of Canonical Discriminant Functions
| Function | Eigenvalue | % of Variance | Cumulative % | Canonical Correlation |
|---|---|---|---|---|
| 1 | 4.513(a) | 67.2 | 67.2 | 0.905 |
| 2 | 2.198(a) | 32.8 | 100.0 | 0.829 |
| a First 2 canonical discriminant functions were used in the analysis. | ||||
Recall that Wilk's lambda is a measure of the discriminating power remaining in the variables, and that values close to 0 indicate high discriminating power. The first value relates to the first function, the second relates to the second function and is measured after removing the discriminating power associated with the first function.
| Test of Function(s) | Wilks' Lambda | Chi-square | df | Sig. |
|---|---|---|---|---|
| 1 through 2 | 0.057 | 96.134 | 16 | 0.000 |
| 2 | 0.313 | 38.945 | 7 | 0.000 |
In this case both functions are significant so both should be retained.
| Function | ||
|---|---|---|
| 1 | 2 | |
| POST | 0.058 | 0.516 |
| PRE | -0.134 | 0.027 |
| BOG | -0.201 | 0.849 |
| CALL | 0.338 | 0.103 |
| WET | 0.866 | -0.063 |
| STEEP | 0.537 | 0.546 |
| LT200 | 0.668 | 1.535 |
| L4_600 | -0.138 | 0.221 |
The Structure Matrix table below shows that:
- function 1 is mainly associated with large areas of wet heath and steep ground, and only small areas of bog (negative correlation). Thus, cases with a positive score on function 1 tend to have more wet heath and steep ground and less bog.
- function 2 is mainly associated with large areas of land below 200m and small areas of steep land, land between 400 & 600m and wet heath. Thus, cases with a positive score on function 2 tend to have more land below 200m, but less steep land and land between 400 & 600 m.
| Function | ||
|---|---|---|
| 1 | 2 | |
| WET | 0.631(*) | -0.428 |
| BOG | -0.467(*) | 0.053 |
| PRE | -0.197(*) | -0.010 |
| LT200 | 0.198 | 0.784(*) |
| STEEP | 0.326 | -0.555(*) |
| L4_600 | -0.037 | -0.443(*) |
| CALL | 0.075 | 0.242(*) |
| POST | -0.077 | 0.194(*) |
| Pooled within-groups correlations between
discriminating variables and standardized canonical discriminant functions Variables ordered by absolute size of correlation within function. |
||
| * Largest absolute correlation between
each variable and any discriminant function |
||
Examining the group centroids allows us to see how the functions separate the groups.
- function 1 separates area 1 from area 2 (-3.2 and +.2.0), with area 3 in between them (-0.4).
- function 2 separates area 3 (+1.6) from the other two (-1.7 and -1.0).
| Function | ||
|---|---|---|
| REGION | 1 | 2 |
| 1 | -3.726 | -1.680 |
| 2 | 2.049 | -1.003 |
| 3 | -0.394 | 1.636 |
| Unstandardized canonical
discriminant functions evaluated at group means |
||
Classification Statistics
The prior probabilities (of class membership) are to be equal, thus they are all 0.333. An alternative weighting would have been to set them to group sizes. For example, this would have given region 1 a prior probability of 0.175 (7/40).
| Prior | Cases Used in Analysis | ||
|---|---|---|---|
| REGION | Unweighted | ||
| 1 | 0.333 | 7 | 7.000 |
| 2 | 0.333 | 16 | 16.000 |
| 3 | 0.333 | 17 | 17.000 |
| Total | 1.000 | 40 | 40.000 |
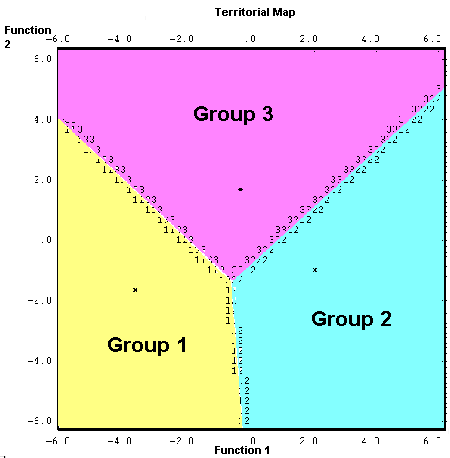
The territorial map (shaded to emphasis groups) highlights how the functions separate the groups (asterisks mark their centroids). Group membership is determined by the combination of function 1 and 2 scores. A coordinate that places a case in the yellow region would indicate a group 1 case.
In the table of case statistics the format is similar to that of the 2 group except that there is an extra column of discriminant function scores and it is possible to have misclassified cases in which even the second highest group is incorrect. [skip table]
| Actual Group |
Highest Group | Second Highest Group | Discriminant Scores | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Predicted Group |
P(G=g | D=d) |
Squared Mahalanobis Distance to Centroid |
Group | P(G=g | D=d) |
Squared Mahalanobis Distance to Centroid |
Function 1 |
Function 2 |
|||||
| Case No. |
P(D>d | G=g) |
|||||||||||
| Original | 1 | 3 | 3 | .620 | 1.000 | .958 | 2 | .000 | 16.778 | -.012 | 2.537 | |
| 2 | 3 | 3 | .231 | .999 | 2.929 | 2 | .001 | 18.112 | .628 | 3.009 | ||
| 3 | 3 | 3 | 1.000 | .998 | .000 | 2 | .002 | 12.922 | -.391 | 1.637 | ||
| 4 | 3 | 3 | .610 | .999 | .989 | 1 | .000 | 16.500 | -1.388 | 1.642 | ||
| 5 | 3 | 3 | .897 | .996 | .217 | 2 | .003 | 11.563 | -.546 | 1.195 | ||
| 6 | 3 | 3 | .263 | .976 | 2.669 | 1 | .024 | 10.086 | -1.839 | .874 | ||
| 7 | 2 | 2 | .860 | 1.000 | .302 | 3 | .000 | 17.150 | 2.369 | -1.450 | ||
| 8 | 2 | 2 | .445 | .996 | 1.619 | 3 | .004 | 12.564 | 2.773 | .043 | ||
| 9 | 2 | 3(**) | .206 | .536 | 3.162 | 2 | .464 | 3.450 | .602 | .162 | ||
| 10 | 2 | 3(**) | .254 | .631 | 2.738 | 2 | .369 | 3.809 | .830 | .522 | ||
| 11 | 2 | 2 | .656 | 1.000 | .845 | 3 | .000 | 19.678 | 2.304 | -1.886 | ||
| 12 | 2 | 2 | .014 | 1.000 | 8.538 | 3 | .000 | 41.981 | 3.525 | -3.525 | ||
| 13 | 2 | 2 | .774 | .999 | .512 | 3 | .001 | 15.465 | 2.723 | -.763 | ||
| 14 | 3 | 3 | .278 | .755 | 2.559 | 2 | .245 | 4.814 | 1.053 | .952 | ||
| 15 | 3 | 3 | .152 | .894 | 3.764 | 1 | .096 | 8.234 | -1.372 | -.040 | ||
| 16 | 3 | 3 | .777 | .998 | .505 | 2 | .001 | 13.774 | -.946 | 1.189 | ||
| 17 | 3 | 3 | .540 | 1.000 | 1.233 | 2 | .000 | 19.349 | -.264 | 2.739 | ||
| 18 | 3 | 3 | .982 | .998 | .036 | 2 | .002 | 12.651 | -.500 | 1.478 | ||
| 19 | 3 | 3 | .211 | .938 | 3.115 | 2 | .043 | 9.291 | -.853 | -.068 | ||
| 20 | 3 | 3 | .707 | .999 | .694 | 2 | .001 | 15.576 | -.001 | 2.370 | ||
| 21 | 3 | 3 | .988 | .999 | .024 | 2 | .001 | 13.414 | -.541 | 1.587 | ||
| 22 | 3 | 3 | .863 | 1.000 | .296 | 2 | .000 | 16.935 | -.619 | 2.130 | ||
| 23 | 3 | 3 | .932 | .996 | .141 | 2 | .004 | 11.062 | -.019 | 1.602 | ||
| 24 | 3 | 3 | .172 | .999 | 3.515 | 2 | .001 | 17.084 | .920 | 2.973 | ||
| 25 | 2 | 2 | .544 | .924 | 1.217 | 3 | .076 | 6.216 | 1.271 | -.221 | ||
| 26 | 2 | 2 | .482 | 1.000 | 1.460 | 3 | .000 | 22.711 | 2.581 | -2.088 | ||
| 27 | 2 | 2 | .108 | .993 | 4.454 | 3 | .005 | 15.227 | .312 | -2.202 | ||
| 28 | 2 | 2 | .887 | 1.000 | .239 | 3 | .000 | 15.920 | 2.086 | -1.490 | ||
| 29 | 2 | 2 | .225 | .590 | 2.982 | 3 | .410 | 3.709 | 1.132 | .461 | ||
| 30 | 2 | 2 | .836 | .988 | .359 | 3 | .012 | 9.220 | 1.811 | -.453 | ||
| 31 | 2 | 2 | .277 | .735 | 2.569 | 3 | .265 | 4.609 | .634 | -.249 | ||
| 32 | 2 | 2 | .121 | 1.000 | 4.224 | 3 | .000 | 24.886 | 4.065 | -.603 | ||
| 33 | 2 | 2 | .099 | 1.000 | 4.627 | 3 | .000 | 32.789 | 3.760 | -2.306 | ||
| 34 | 1 | 1 | .555 | 1.000 | 1.177 | 3 | .000 | 33.372 | -4.599 | -2.325 | ||
| 35 | 1 | 1 | .355 | .992 | 2.070 | 3 | .008 | 11.764 | -2.387 | -1.156 | ||
| 36 | 1 | 1 | .789 | 1.000 | .475 | 3 | .000 | 29.054 | -4.221 | -2.160 | ||
| 37 | 1 | 1 | .615 | .999 | .972 | 3 | .001 | 15.169 | -2.767 | -1.452 | ||
| 38 | 1 | 1 | .673 | 1.000 | .792 | 3 | .000 | 23.128 | -4.372 | -1.067 | ||
| 39 | 1 | 1 | .514 | .996 | 1.331 | 3 | .004 | 12.633 | -2.834 | -.948 | ||
| 40 | 1 | 1 | .311 | 1.000 | 2.334 | 3 | .000 | 38.734 | -4.906 | -2.651 | ||
| Cross- validated (a) |
1 | 3 | 3 | .817 | 1.000 | 4.429 | 2 | .000 | 19.707 | |||
| 2 | 3 | 3 | .121 | .999 | 12.737 | 2 | .001 | 26.834 | ||||
| 3 | 3 | 3 | .959 | .998 | 2.550 | 2 | .002 | 14.832 | ||||
| 4 | 3 | 3 | .001 | .981 | 27.488 | 1 | .019 | 35.355 | ||||
| 5 | 3 | 3 | .999 | .996 | .878 | 2 | .004 | 11.800 | ||||
| 6 | 3 | 3 | .235 | .882 | 10.453 | 1 | .117 | 14.490 | ||||
| 7 | 2 | 2 | .868 | 1.000 | 3.880 | 3 | .000 | 20.306 | ||||
| 8 | 2 | 2 | .001 | .959 | 25.752 | 3 | .041 | 32.037 | ||||
| 9 | 2 | 3(**) | .669 | .789 | 5.808 | 2 | .211 | 8.444 | ||||
| 10 | 2 | 3(**) | .024 | .999 | 17.627 | 2 | .001 | 30.883 | ||||
| 11 | 2 | 2 | .136 | 1.000 | 12.354 | 3 | .000 | 31.355 | ||||
| 12 | 2 | 2 | .007 | 1.000 | 21.157 | 3 | .000 | 66.711 | ||||
| 13 | 2 | 2 | .971 | .999 | 2.285 | 3 | .001 | 16.706 | ||||
| 14 | 3 | 2(**) | .146 | .756 | 12.121 | 3 | .244 | 14.377 | ||||
| 15 | 3 | 3 | .386 | .735 | 8.501 | 1 | .242 | 10.721 | ||||
| 16 | 3 | 3 | .164 | .995 | 11.724 | 2 | .003 | 23.399 | ||||
| 17 | 3 | 3 | .432 | 1.000 | 8.010 | 2 | .000 | 26.007 | ||||
| 18 | 3 | 3 | .998 | .998 | 1.081 | 2 | .002 | 13.224 | ||||
| 19 | 3 | 3 | .400 | .829 | 8.350 | 2 | .112 | 12.360 | ||||
| 20 | 3 | 3 | .707 | .999 | 5.466 | 2 | .001 | 19.625 | ||||
| 21 | 3 | 3 | .670 | .998 | 5.799 | 2 | .002 | 18.235 | ||||
| 22 | 3 | 3 | .897 | 1.000 | 3.528 | 2 | .000 | 19.754 | ||||
| 23 | 3 | 3 | .036 | .981 | 16.490 | 2 | .019 | 24.432 | ||||
| 24 | 3 | 3 | .235 | .998 | 10.442 | 2 | .002 | 22.626 | ||||
| 25 | 2 | 2 | .759 | .867 | 4.987 | 3 | .133 | 8.744 | ||||
| 26 | 2 | 2 | .247 | 1.000 | 10.269 | 3 | .000 | 32.484 | ||||
| 27 | 2 | 1(**) | .000 | .911 | 29.269 | 2 | .080 | 34.122 | ||||
| 28 | 2 | 2 | .526 | .999 | 7.102 | 3 | .001 | 22.058 | ||||
| 29 | 2 | 3(**) | .040 | .982 | 16.159 | 2 | .018 | 24.136 | ||||
| 30 | 2 | 2 | .251 | .962 | 10.206 | 3 | .038 | 16.687 | ||||
| 31 | 2 | 3(**) | .044 | .925 | 15.889 | 2 | .075 | 20.927 | ||||
| 32 | 2 | 2 | .623 | 1.000 | 6.216 | 3 | .000 | 27.273 | ||||
| 33 | 2 | 2 | .114 | 1.000 | 12.949 | 3 | .000 | 45.958 | ||||
| 34 | 1 | 1 | .757 | 1.000 | 5.003 | 3 | .000 | 37.045 | ||||
| 35 | 1 | 1 | .124 | .877 | 12.675 | 3 | .121 | 16.639 | ||||
| 36 | 1 | 1 | .980 | 1.000 | 2.018 | 3 | .000 | 29.929 | ||||
| 37 | 1 | 1 | .275 | .993 | 9.865 | 3 | .007 | 19.847 | ||||
| 38 | 1 | 3(**) | .000 | .747 | 89.792 | 1 | .253 | 91.955 | ||||
| 39 | 1 | 1 | .784 | .992 | 4.747 | 3 | .008 | 14.292 | ||||
| 40 | 1 | 1 | .124 | 1.000 | 12.654 | 3 | .000 | 50.946 | ||||
| For the original data, squared
Mahalanobis distance is based on canonical functions. For the cross-validated data, squared Mahalanobis distance is based on observations. |
||||||||||||
| ** Misclassified case | ||||||||||||
| a Cross validation is done
only for those cases in the analysis. In cross validation, each case is classified by the functions derived from all cases other than that case. |
||||||||||||
Again the results are summarised in confusion matrices, this time 3 x 3 because there are 3 groups.
| Predicted Group Membership | Total | |||||
|---|---|---|---|---|---|---|
| Region | 1 | 2 | 3 | |||
| Original | Count | 1 | 7 | 0 | 0 | 7 |
| 2 | 0 | 14 | 2 | 16 | ||
| 3 | 0 | 0 | 17 | 17 | ||
| % | 1 | 100.0 | .0 | .0 | 100.0 | |
| 2 | .0 | 87.5 | 12.5 | 100.0 | ||
| 3 | .0 | .0 | 100.0 | 100.0 | ||
| Cross-validated(a) | Count | 1 | 6 | 0 | 1 | 7 |
| 2 | 1 | 11 | 4 | 16 | ||
| 3 | 0 | 1 | 16 | 17 | ||
| % | 1 | 85.7 | .0 | 14.3 | 100.0 | |
| 2 | 6.3 | 68.8 | 25.0 | 100.0 | ||
| 3 | .0 | 5.9 | 94.1 | 100.0 | ||
| a Cross validation is done only for those
cases in the analysis. In cross validation, each case is classified by the functions derived from all cases other than that case. |
||||||
| b 95.0% of original grouped cases correctly classified. | ||||||
| c 82.5% of cross-validated grouped cases correctly classified. | ||||||
The regions are very accurately predicted using the resubstitution (original) method, only two region 2 cases are misclassified. Even using the cross-validated method accuracy remains good, although five region 2 cases are now misclassified.
The results are shown graphically. Axes are the the two discriminant functions and the coordinates are the scores on the two axes. Regions are colour coded.
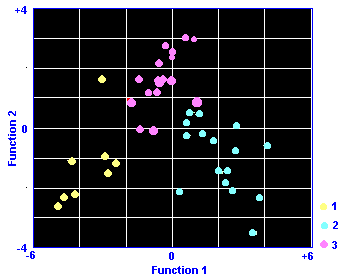
In summary
- Using these habitat variables we can discriminate between the core area habitats of golden eagles living in 3 Scottish regions.
- We have identified how the habitats differ, two gradients were detected.
However, rather a large number of variables (8) were used with a relatively small number of cases (40). Such ratios tend to give very good separation. Various ratios have been suggested in the literature. The range of n:p is between 3:1 and 5:1, where n is the smallest group size and p is the number of predictors. The smallest group size was seven suggesting that no more than 2 predictors should be used.
The next analysis uses a stepwise analysis in an attempt to reduce the predictor dimensionality.


