Duchenne Muscular Dystrophy: Discriminant Analysis
All Variables used
The following output was obtained using SPSS.
| Duchenne Muscular Dystrophy | Mean | SD | n | ||
|---|---|---|---|---|---|
| NonCarrier | creatine kinase | 43.051 | 22.206 | 39 | |
| hemopexin | 79.615 | 12.046 | 39 | ||
| lactate dehydrogenase | 12.536 | 4.788 | 39 | ||
| pyruvate kinase | 164.974 | 39.846 | 39 | ||
| Carrier | creatine kinase | 155.618 | 159.854 | 34 | |
| hemopexin | 94.009 | 11.220 | 34 | ||
| lactate dehydrogenase | 26.271 | 20.665 | 34 | ||
| pyruvate kinase | 247.500 | 67.762 | 34 | ||
| Total | creatine kinase | 95.479 | 123.162 | 73 | |
| hemopexin | 86.319 | 13.658 | 73 | ||
| lactate dehydrogenase | 18.933 | 15.982 | 73 | ||
| pyruvate kinase | 203.411 | 68.269 | 73 | ||
Comparing class means
Which predictors can the two classes be separated on?
In fact, using an F-test (ANOVA), they differ with repect to all 4 variables. The F statistics are a guide to the extent (reliability) of the differences between the classes for each variable. Using the F values as guide, make a note of the rank order of the predictor variables.
| Wilks' Lambda | F | Sig. | |
|---|---|---|---|
| creatine kinase | 0.789 | 18.96 | 0.000 |
| hemopexin | 0.720 | 27.63 | 0.000 |
| lactate dehydrogenase | 0.814 | 16.26 | 0.000 |
| pyruvate kinase | 0.631 | 41.46 | 0.000 |
Wilk's lambda is a multivariate test statistic whose value ranges between 0 and 1. Values close to 0 indicate that the class means are different and values close to 1 indicating the class means are not different (equal to 1 indicates all means are the same).
Correlations and covariances
The following table is presented for information only. It is not normally shown in an SPSS analysis. However, the correlation matrix is useful because it allows us to examine the discriminating variables for evidence of collinearity (correlation - which implies a certain redundancy in the discriminating variables). This can cause problems similar to those observed in multiple regression.
| creatine kinase |
hemopexin | lactate dehydrogenase |
pyruvate kinase |
||
|---|---|---|---|---|---|
| Covariance | creatine kinase | 12140.760 | |||
| hemopexin | -35.807 | 136.176 | |||
| lactate dehydrogenase | 1244.181 | -2.795 | 210.761 | ||
| pyruvate kinase | 2273.458 | 46.802 | 393.112 | 2983.936 | |
| Correlation | creatine kinase | 1.000 | |||
| hemopexin | -0.028 | 1.000 | |||
| lactate dehydrogenase | 0.778 | -0.017 | 1.000 | ||
| pyruvate kinase | 0.378 | 0.073 | 0.496 | 1.000 | |
Hemopexin does not appear to be correlated with the other 3 variables, which appear to be related, especially creatine and lactate dehydrogenase. Remember these correlation patterns, they will be relevant later.
Again the next tables are presented for information. An assumption of discriminant analysis is that there is no evidence of a difference between the covariance matrices of the two classs. There are formal significance tests (e.g. Box's M) but they are not very robust. In particular they are generally thought to be too powerful, i.e the null hypothesis is rejected even when there are minor differences, Box's M is also susceptible to deviations from multivariate normality (another assumprion). If Box's test is applied to these data we would conclude that the covariance matrices were not equal. This is not too surprising given the differences, e.g. 493.1 cf 25553.2 and 7.927 & 2667.746.
| Duchenne Muscular Dystrophy | creatine kinase |
hemopexin | lactate dehydrogenase |
pyruvate kinase |
|
|---|---|---|---|---|---|
| NonCarrier | creatine kinase | 493.103 | |||
| hemopexin | -44.172 | 145.110 | |||
| lactate dehydrogenase | 7.927 | 9.222 | 22.921 | ||
| pyruvate kinase | -6.683 | 141.737 | 70.898 | 1587.710 | |
| Carrier | creatine kinase | 25553.213 | |||
| hemopexin | -26.175 | 125.889 | |||
| lactate dehydrogenase | 2667.746 | -16.633 | 427.061 | ||
| pyruvate kinase | 4899.076 | -62.517 | 764.145 | 4591.712 | |
| Total | creatine kinase | 15168.864 | |||
| hemopexin | 373.443 | 186.551 | |||
| lactate dehydrogenase | 1616.947 | 47.117 | 255.424 | ||
| pyruvate kinase | 4585.495 | 345.821 | 673.606 | 4660.662 | |
Summary of Canonical Discriminant Functions
| Function | Eigenvalue | % of Variance | Cumulative % | Canonical Correlation |
|---|---|---|---|---|
| 1 | 0.994(a) | 100.0 | 100.0 | 0.706 |
| a First 1 canonical discriminant functions were used in the analysis. | ||||
The canonical correlation is the square root of the ratio of the between-groups sum of squares to the total sum of squares. Squared, it is the proportion of the total variability explained by differences between classs. Thus, if all of the variability in the variables was a consequence of the class differences the canonical correlation would be 1, while if none of the variability was due to class differences the canonical correlation would be 0.
| Test of Function(s) | Wilks' Lambda | Chi-square | df | Sig. |
|---|---|---|---|---|
| 1 | 0.502 | 47.617 | 4 | 0.000 |
Recall that Wilk's lambda measures differences between classs. It can be converted into a Chi-square statistic so that a significance test can be applied. The null hypothesis to be tested is that 'There is no discriminating power remaining in the variables'. Since p is less than 0.001 we would normally reject Ho. This implies that the variables have some ability to discriminate between the classs.
Note that if p had been >0.05 we would normally halt the analysis since there would be no evidence that our variables were able to discriminate between the classs.
The next table give the standardised values for the coefficients. In other words they are weights that would be applied to standardised variables in order to calculate the discriminant scores.
| Function | |
|---|---|
| 1 | |
| creatine kinase | 0.402 |
| hemopexin | 0.588 |
| lactate dehydrogenase | -0.141 |
| pyruvate kinase | 0.641 |
The predictors can be ranked using these standardised variables (ignoring the sign). This implies that pyruvate kinase is the 'best' discriminator (because its weight indicates that it contributes the most to the discriminant score) and that lactate dehydrogenase is the worst.
Discriminant Score = 0.402.creatine + 0.588.hemopexin - 0.141.lactate + 0.641.pyruvate
Although the coefficients can be used for interpretitive purposes there are problems when, as in this case, there is correlation between the predictors. The weights reflect the contribution made by a varaible, after accounting for the discrimination achieved using other variables. Consider an extreme example of two perfectly correlated predictors (A and B). If A has already been used to discrimate then B cannot provide any extra discrimination (even though on its own it is just as good as A).
The structure matrix is a better index of class differences.
| Function | |
|---|---|
| 1 | |
| pyruvate kinase | 0.766 |
| hemopexin | 0.626 |
| creatine kinase | 0.518 |
| lactate dehydrogenase | 0.480 |
| Pooled within-groups correlations
between discriminating variables and standardized canonical discriminant functions Variables ordered by absolute size of correlation within function. |
|
These function values are simply the correlation coefficients between the discriminant score and the predictor variable scores. Note how the strongest correlation is with the pyruvate value. These correlations are shown graphically below.
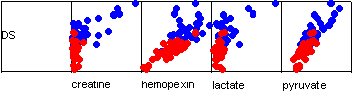
The problem with standardised coefficients is that it is difficult to apply the function to new data. One solution is to obtain unstandardised coefficients. Note that it is not easy to rank these unstandardised coefficients since their magnitudes are related to the scale of their associated predictor.
| Function | |
|---|---|
| 1 | |
| creatine kinase | 0.004 |
| hemopexin | 0.050 |
| lactate dehydrogenase | -0.010 |
| pyruvate kinase | 0.012 |
| (Constant) | -6.899 |
Using these values we can construct a discriminant function that can be used to determine a Discriminant Score using raw variables.
Discriminant Score = -6.899 + 0.004.creatine + 0.050.hemopexin - 0.010.lactate + 0.012.pyruvate
The mean discriminant scores for the two classs are shown below.
| Function | |
|---|---|
| Duchenne Muscular Dystrophy | 1 |
| NonCarrier | -0.918 |
| Carrier | 1.053 |
Classification Statistics
How well does our discriminant function perform? Allocation of cases to classs is based on a probability calculation that uses Bayesian methods. Part of this calculation demands a knowledge of the prior probabilities, i.e. the probability that a case will belong to particular class in the absence of any other discriminating information. The default calculation assumes an equal probability for each class. In some circumstances they may be unequal, for example if members of one class are very rare then we might expect that class's prior probability to be lower than other classs.
| Prior | Cases | ||
|---|---|---|---|
| Duchenne Muscular Dystrophy | |||
| NonCarrier | 0.500 | 39 | |
| Carrier | 0.500 | 34 | |
| Total | 1.000 | 73 | |
The following are presented for historical purposes. The original method of discriminating between classs was developed by Fisher. In his approach a separate function is derived for each class.
| Duchenne Muscular Dystrophy | ||
|---|---|---|
| NonCarrier | Carrier | |
| creatine kinase | -0.004 | 0.003 |
| hemopexin | 0.567 | 0.666 |
| lactate dehydrogenase | -0.004 | -0.023 |
| pyruvate kinase | 0.050 | 0.073 |
| (Constant) | -27.243 | -40.974 |
The next table contains a lot of information. Firstly, the table rows can be split into two main components:
- Original (resubstitution of original cases into the discriminant function)
- Cross-validated (The discriminant function is recalculated using all but the current case - the resultant discriminant function is applied to the missing case and a prediction is made. Because the true and predicted class of each case is known a reasonably 'independent measure of the function's predictive power is obtained).
Next the rows. Some are obvious such as case number and actual class. The final column is the discriminat socre for a case. Consider case 1, which has a score of -0.544. The understandardised discriminant function could be applied to the data from the case.
| variable x |
coefficient (weight) |
value | w.x |
|---|---|---|---|
| creatine | 0.004 | 52.0 | 0.208 |
| hemopexin | 0.050 | 83.5 | 4.175 |
| lactate | -0.013 | 10.9 | -0.142 |
| pyruvate | 0.012 | 176.0 | 2.112 |
| constant | -6.899 | -6.899 | |
| Sum | -0.546 |
The remaining rows are split into two sections
- Highest group (the group which is the most probable, given the discriminant score).
- Second highest group (the group with the next highest probability, given the discriminant score).
There are now three columns to consider:
| P(D>d | G=g) | This is the probability of the discriminant score given the predicted class, you can think of this as a type of z test. |
|---|---|
| P(G=g | D=d) | P(G/D) is the probability of the group given the discriminant
score, hence it is the probability that a case belongs to the predicted
class. If the predicted and actual classs are not the same the case is marked ** (for example case 4). P(G/D) is also given for the second most probable class. Note that the probability cutoff is very strict. If P(carrier) = 0.501 and P(non-carrier) = 0.499 the individual would be classified as a carrier. It is important that you examine the probabilities for misclassifed cases to determine if they are a serious or a marginal misclassification. Misclassified individuals can be useful, if you can find out why they were misclassified it may tell you a lot about the nature of the normal differences between classs. For example, suppose you are discriminating between 'normal' and diseased individuals and some normals are predicted to be diseased. What is special about these individuals (possibly other features not included in the analysis) that has kept them healthy? |
| Squared Mahalanobis Distance to Centroid |
A measure of how much a case's values differ from the average of all cases in the class. For a single variable, it is simply the square of the standardized value of the independent variable (because the mean will be 0 for a standardised varaible). A large Mahalanobis distance identifies a case as having extreme values on one or more of the independent variables. Thus, it measures how far a case is from the 'mean' of its class. Note that as this increases the probability of belonging to a class decreases. |
| Actual class |
Highest class | Second Highest class | Discriminant Scores |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Predicted class |
P(D>d | G=g) | P(G=g | D=d) | Squared Mahalanobis Distance to Centroid |
class | P(G=g | D=d) | Squared Mahalanobis Distance to Centroid |
|||||
| Case No. |
p | df | |||||||||
| Original | 1 | 1 | 1 | .709 | 1 | .770 | .140 | 2 | .230 | 2.552 | -.544 |
| 2 | 1 | 1 | .833 | 1 | .822 | .044 | 2 | .178 | 3.100 | -.708 | |
| 3 | 1 | 1 | .722 | 1 | .776 | .127 | 2 | .224 | 2.608 | -.562 | |
| 4 | 1 | 2(**) | .900 | 1 | .845 | .016 | 1 | .155 | 3.407 | .928 | |
| 5 | 1 | 1 | .546 | 1 | .680 | .364 | 2 | .320 | 1.871 | -.315 | |
| 6 | 1 | 1 | .806 | 1 | .919 | .061 | 2 | .081 | 4.916 | -1.164 | |
| 7 | 1 | 2(**) | .355 | 1 | .530 | .854 | 1 | .470 | 1.096 | .129 | |
| 9 | 1 | 1 | .902 | 1 | .899 | .015 | 2 | .101 | 4.384 | -1.041 | |
| 39 | 1 | 1 | .859 | 1 | .831 | .031 | 2 | .169 | 3.218 | -.741 | |
| 40 | 2 | 2 | .245 | 1 | .986 | 1.354 | 1 | .014 | 9.826 | 2.217 | |
| 41 | 2 | 2 | .380 | 1 | .553 | .771 | 1 | .447 | 1.195 | .175 | |
| 42 | 2 | 2 | .399 | 1 | .974 | .713 | 1 | .026 | 7.926 | 1.897 | |
| 43 | 2 | 2 | .481 | 1 | .635 | .496 | 1 | .365 | 1.604 | .348 | |
| 44 | 2 | 1(**) | .465 | 1 | .623 | .533 | 2 | .377 | 1.539 | -.188 | |
| 45 | 2 | 2 | .099 | 1 | .994 | 2.726 | 1 | .006 | 13.119 | 2.704 | |
| 46 | 2 | 2 | .715 | 1 | .772 | .134 | 1 | .228 | 2.578 | .688 | |
| 47 | 2 | 2 | .445 | 1 | .607 | .584 | 1 | .393 | 1.457 | .289 | |
| 48 | 2 | 2 | .746 | 1 | .787 | .105 | 1 | .213 | 2.715 | .730 | |
| 49 | 2 | 2 | .863 | 1 | .832 | .030 | 1 | .168 | 3.234 | .880 | |
| 50 | 2 | 1(**) | .528 | 1 | .668 | .398 | 2 | .332 | 1.796 | -.287 | |
| 70 | 2 | 2 | .446 | 1 | .608 | .582 | 1 | .392 | 1.460 | .290 | |
| 71 | 2 | 2 | .540 | 1 | .959 | .375 | 1 | .041 | 6.675 | 1.666 | |
| 72 | 2 | 2 | .772 | 1 | .798 | .084 | 1 | .202 | 2.828 | .764 | |
| Cross- validated(a) |
1 | 1 | 1 | .991 | 4 | .765 | .283 | 2 | .235 | 2.649 | |
| 2 | 1 | 1 | .936 | 4 | .815 | .819 | 2 | .185 | 3.783 | ||
| 3 | 1 | 1 | .979 | 4 | .771 | .439 | 2 | .229 | 2.862 | ||
| 4 | 1 | 2(**) | .529 | 4 | .893 | 3.172 | 1 | .107 | 7.425 | ||
| 5 | 1 | 1 | .948 | 4 | .673 | .730 | 2 | .327 | 2.175 | ||
| 6 | 1 | 1 | .999 | 4 | .916 | .080 | 2 | .084 | 4.867 | ||
| 7 | 1 | 2(**) | .719 | 4 | .556 | 2.089 | 1 | .444 | 2.537 | ||
| 8 | 1 | 1 | .763 | 4 | .555 | 1.850 | 2 | .445 | 2.294 | ||
| 9 | 1 | 1 | .547 | 4 | .890 | 3.063 | 2 | .110 | 7.237 | ||
| 39 | 1 | 1 | .833 | 4 | .822 | 1.463 | 2 | .178 | 4.519 | ||
| 40 | 2 | 2 | .102 | 4 | .986 | 7.722 | 1 | .014 | 16.195 | ||
| 41 | 2 | 2 | .725 | 4 | .531 | 2.058 | 1 | .469 | 2.304 | ||
| 42 | 2 | 2 | .255 | 4 | .972 | 5.332 | 1 | .028 | 12.419 | ||
| 43 | 2 | 2 | .413 | 4 | .592 | 3.951 | 1 | .408 | 4.693 | ||
| 44 | 2 | 1(**) | .839 | 4 | .650 | 1.427 | 2 | .350 | 2.663 | ||
| 45 | 2 | 2 | .000 | 4 | .997 | 36.218 | 1 | .003 | 47.815 | ||
| 46 | 2 | 2 | .670 | 4 | .753 | 2.362 | 1 | .247 | 4.596 | ||
| 47 | 2 | 2 | .853 | 4 | .594 | 1.350 | 1 | .406 | 2.110 | ||
| 48 | 2 | 2 | .877 | 4 | .776 | 1.209 | 1 | .224 | 3.697 | ||
| 49 | 2 | 2 | .953 | 4 | .826 | .688 | 1 | .174 | 3.802 | ||
| 50 | 2 | 1(**) | .038 | 4 | .815 | 10.147 | 2 | .185 | 13.116 | ||
| 59 | 2 | 2 | .002 | 4 | .999 | 16.637 | 1 | .001 | 30.962 | ||
| 72 | 2 | 2 | .693 | 4 | .781 | 2.235 | 1 | .219 | 4.781 | ||
| 73 | 2 | 2 | .306 | 4 | .792 | 4.819 | 1 | .208 | 7.494 | ||
| For the original
data, squared Mahalanobis distance is based on canonical functions. For the cross-validated data, squared Mahalanobis distance is based on observations. |
|||||||||||
| ** Misclassified case | |||||||||||
| a Cross validation
is done only for those cases in the analysis. In cross validation, each case is classified by the functions derived from all cases other than that case. |
|||||||||||
Although the case listing can be useful it becomes less useful as the number of cases increases. Often all that is required is a summary of the performance. In the following confusion matrices results are shown for resubstituted (original) data and the more robust cross-validated results. It is the second of these that indicate how well the function is likely to generalise to new cases. The performance is almost always worse using resubstituted data. More information about this can be found in the accuracy pages.
| Predicted class Membership |
Total | ||||
|---|---|---|---|---|---|
| Duchenne Muscular Dystrophy |
NonCarrier | Carrier | |||
| Original | Count | NonCarrier | 36 | 3 | 39 |
| Carrier | 6 | 28 | 34 | ||
| % | NonCarrier | 92.3 | 7.7 | 100.0 | |
| Carrier | 17.6 | 82.4 | 100.0 | ||
| Cross- validated(a) |
Count | NonCarrier | 35 | 4 | 39 |
| Carrier | 6 | 28 | 34 | ||
| % | NonCarrier | 89.7 | 10.3 | 100.0 | |
| Carrier | 17.6 | 82.4 | 100.0 | ||
| a Cross validation is done
only for those cases in the analysis. In cross validation, each case is classified by the functions derived from all cases other than that case. |
|||||
| b 87.7% of original grouped cases correctly classified. | |||||
| c 86.3% of cross-validated grouped cases correctly classified. | |||||
This discriminant function appears to have performed quite well, with about 87% of cases correctly classified. Using the cross-validated data 4 non-carriers (10.3%) were classified as carriers and 6 carriers (17.6%) were classified as carriers. Although this may seem quite good we have to question if this level of inaccuracy, particularly the misclassified carriers, would be acceptable in practice. The accuracy pages deal, in greater depth, with the problem of measuring classification accuracy.


