Golden Eagle Core Areas: User pruning of predictors
We are allowed to use some judgement in the selection of predictors. Indeed we should be encouraged to do so since it means that we are thinking more deeply about the problem. Huberty (1994) suggests that we should use three variable screening techniques:
Logical screening:
Uses theoretical, reliability and practical grounds to screen variables. Do some initial research to find variables that may have some theoretical link with our groupings. You may also wish to take into account the data reliability. Finally do not ignore the practical problems of obtaining data, this includes cost (time or financial) factors.
Statistical screening:
Uses statistical tests to identify variables whose values differ significantly between groups. Huberty suggests applying a relaxed criterion, in that we only reject those variables that are most likely to be 'noise' (e.g. F <1.0 or t < 1.0). In addition, examine the inter-predictor correlations to find those variables that are 'highly correlated'. Redundant variables should be removed.
Dimension reduction:
Use a method such as PCA to reduce the predictor dimensionality. This has the additional advantage of removing any predictor collinearity.
This analysis concentrates on statistical screening and applies rather stricter criteria than Huberty suggests. This is because of the need to drastically prune the number of variables. The first step is to carry out a single factor analysis of varaince to find out which variables discriminate between the regions. The F statistics are used to rank the potential predictors. Non-significant predictors (POST< PRE and CAL) have p values <0.05. Note that if a Bonferroni correction for multiple testing is applied the p value of 0.05 for PRE becomes insignificant.
| SS | df | MS | F | Sig. | ||
|---|---|---|---|---|---|---|
| POST | Between Groups | 26.569 | 2 | 13.284 | 2.022 | .147 |
| Within Groups | 243.059 | 37 | 6.569 | |||
| Total | 269.628 | 39 | ||||
| PRE | Between Groups | 40.811 | 2 | 20.406 | 3.251 | .050 |
| Within Groups | 232.213 | 37 | 6.276 | |||
| Total | 273.024 | 39 | ||||
| BOG | Between Groups | 386.218 | 2 | 193.109 | 18.360 | .000 |
| Within Groups | 389.152 | 37 | 10.518 | |||
| Total | 775.370 | 39 | ||||
| CALL | Between Groups | 24.370 | 2 | 12.185 | 2.845 | .071 |
| Within Groups | 158.466 | 37 | 4.283 | |||
| Total | 182.836 | 39 | ||||
| WET | Between Groups | 393.277 | 2 | 196.638 | 40.687 | .000 |
| Within Groups | 178.819 | 37 | 4.833 | |||
| Total | 572.096 | 39 | ||||
| STEEP | Between Groups | 461.580 | 2 | 230.790 | 21.367 | .000 |
| Within Groups | 399.651 | 37 | 10.801 | |||
| Total | 861.231 | 39 | ||||
| LT200 | Between Groups | 1265.152 | 2 | 632.576 | 28.273 | .000 |
| Within Groups | 827.832 | 37 | 22.374 | |||
| Total | 2092.984 | 39 | ||||
| L4_600 | Between Groups | 139.423 | 2 | 69.711 | 8.078 | .001 |
| Within Groups | 319.296 | 37 | 8.630 | |||
| Total | 458.719 | 39 | ||||
The rank order (based on F statistics) is:
- Wet (40.687);
- LT200 (28.273);
- Steep (21.367);
- Bog (18.360);
- L4_600 (8.078).
The next analysis is restricted to these 5 predictors.
Do they convey independent information, or are some of them too correlated?
| WET | LT200 | STEEP | BOG | ||
|---|---|---|---|---|---|
| LT200 | Pearson Correlation | -.203 | 1.000 | ||
| Sig. (2-tailed) | .209 | . | |||
| STEEP | Pearson Correlation | .690 | -.547 | 1.000 | |
| Sig. (2-tailed) | .000 | .000 | . | ||
| BOG | Pearson Correlation | -.543 | -.314 | -.462 | 1.000 |
| Sig. (2-tailed) | .000 | .049 | .003 | . | |
| L4_600 | Pearson Correlation | .266 | -.700 | .592 | -.058 |
| Sig. (2-tailed) | .097 | .000 | .000 | .722 | |
It is apparent from this table that LT200 & L4_600 are highly correlated (r = -0.700). Since LT200 has the larger F statistic we will exclude L4_600.
Similarly WET & STEEP are highly correlated (r = 0.690). For similar reasons STEEP is excluded.
This leaves WET, LT200 & BOG.
BOG is reasonably correlated with the other two (-0.543 with BOG and -0.314 with LT200), while WET and LT200 have an insignificant correlation (r = -0.203, p = 0.209).
Therefore, exclude BOG and retain only WET and LT200. Now there are the desired 2 predictors (to fulfil the desired cases:predictors ratio). These are now used in a discriminant analysis. (Note that these two predictors were the most important in the previous two analyses.)
Summary of Canonical Discriminant Functions
As in the previous analyses we extract two discriminant functions.
| Function | Eigenvalue | % of Variance | Cumulative % | Canonical Correlation |
|---|---|---|---|---|
| 1 | 2.930 | 68.3 | 68.3 | 0.863 |
| 2 | 1.359 | 31.7 | 100.0 | 0.759 |
| Test of Function(s) | Wilks' Lambda | Chi-square | df | Sig. |
|---|---|---|---|---|
| 1 through 2 | .108 | 81.286 | 4 | .000 |
| 2 | .424 | 31.327 | 1 | .000 |
| Function | ||
|---|---|---|
| 1 | 2 | |
| WET | 1.033 | -0.359 |
| LT200 | 0.746 | 0.800 |
| Function | ||
|---|---|---|
| 1 | 2 | |
| WET | 0.731 | -0.682 |
| LT200 | 0.328 | 0.945 |
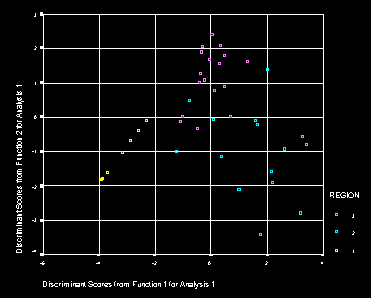
The structure matrix suggests that the first function is primarly associated with wet heath (WET). Cases with a high positive score have more wet heath. The second function comprises both WET & LT200. Larger positive scores are associated with land that has less wet heath and more land below 200m.
As in the other analyses function 1 separates regions 1 and 2, while function 2 separates region 3 from the rest.
| Function | ||
|---|---|---|
| REGION | 1 | 2 |
| 1 | -3.220 | -1.058 |
| 2 | 1.495 | -0.922 |
| 3 | -0.081 | 1.303 |
Classification Statistics
Prior probabilities are set to equal (0.333 for each region).
| Predicted Group Membership | Total | |||||
|---|---|---|---|---|---|---|
| REGION | 1 | 2 | 3 | |||
| Original | Count | 1 | 7 | 0 | 0 | 7 |
| 2 | 1 | 12 | 3 | 16 | ||
| 3 | 0 | 0 | 17 | 17 | ||
| % | 1 | 100.0 | .0 | .0 | 100.0 | |
| 2 | 6.3 | 75.0 | 18.8 | 100.0 | ||
| 3 | .0 | .0 | 100.0 | 100.0 | ||
| Cross-validated | Count | 1 | 7 | 0 | 0 | 7 |
| 2 | 1 | 12 | 3 | 16 | ||
| 3 | 0 | 0 | 17 | 17 | ||
| % | 1 | 100.0 | .0 | .0 | 100.0 | |
| 2 | 6.3 | 75.0 | 18.8 | 100.0 | ||
| 3 | .0 | .0 | 100.0 | 100.0 | ||
This analysis, based on only two predictors, produces a better discrimination than the one using all 8. It is only marginally worse than the stepwise analysis with 3 predictors (Cross-validated results: region 2 has 1 newly misclassified as region 1 and 1 extra region 3 misclassification).